ICTV Computational Virus Taxonomy Challenge

A community challenge to evaluate and compare viral genome taxonomy classifiers
Why do we need a computational taxonomy?
There is a consensus that viruses are so diverse that no single taxonomic method can be used to classify them all (Simmonds et al. PLoS Biology 2023). Since its inception, the ICTV has been seeking the expertise and domain-specific knowledge of the global virology community to classify viruses. This has generated a patchwork of methods that, ideally, capture the features of different viral lineages and generate meaningful taxa that are in agreement with biology. These methods are formalized in taxonomy proposals (TaxoProps) written by experts and ratified by the ICTV. These documents describe how viruses within each taxon shall be classified, and include specific demarcation criteria. The TaxoProps are available as Word documents on the ICTV website.
As metagenomics is rapidly expanding our view of the virosphere, we are looking to make sense of the sequences we discover. The number and diversity of sequences found in viromics datasets is staggering and make their taxonomic classification a daunting task that one would ideally approach using computational tools. There is currently no ICTV-approved method to approach this question. While the solution will likely not be trivial, we have to face this challenge to keep up with the growth of viruses that we aim to classify. The ICTV Computational Virus Taxonomy Challenge is our first attempt at assessing the different computational tools that are available for virus taxonomy.
The challenge
The ICTV Computational Virus Taxonomy Challenge is a challenge for bioinformaticians who build methods to classify unknown sequences into the current ICTV-approved taxonomy. We ask you to classify a set of viral sequences using a bioinformatics pipeline of your choice or design. The classification will be evaluated using the Taxonomy Release MSL39. Importantly, we ask that your pipeline be fully and easily reproducible and that you make available the necessary code and environment to run it. Multiple strategies exist, such as the creation of virtual environments (venv, conda) or containers (Singularity, Docker), and the code should be made available via GitHub. The results of your classification will be further evaluated on metrics including reproducability, speed, accuracy at different ranks, for different types of viruses, etc.
In practice
For the challenge, we have collected thousands of viral sequences that experts have classified into the various ranks of the ICTV taxonomy.
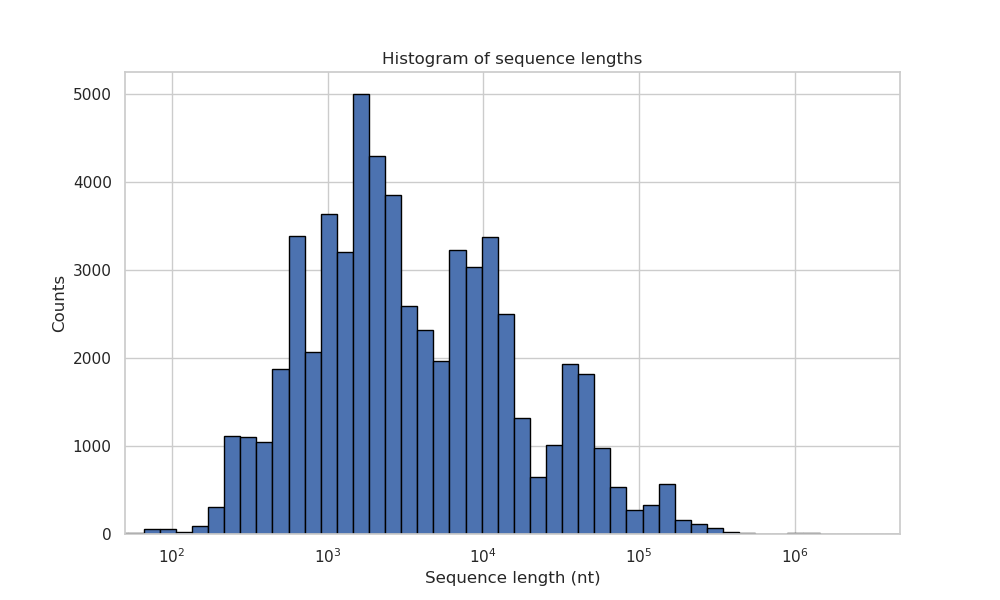 Sequence length distribution of the contigs in the ICTV Computational Virus Taxonomy Challenge.
Sequence length distribution of the contigs in the ICTV Computational Virus Taxonomy Challenge.
The fasta sequences are available in the compressed folder here. Every sequence in the folder is a different virus contig or genome fragment with unknown accuracy and unknown completeness. The idea is that these sequences might have resulted from a metagenomics experiment, and your challenge is to classify them into ICTV-approved taxa at the lowest appropriate rank. For example, if the sequence represents a new species in a known genus, it should be annotated down to the genus rank, but not below. We ask that your pipeline returns a tab-separated values (.csv) file where each rows includes the contig header (TaxoChallenge_number) and 31 the columns listed below. Fields can be left empty if no annotation is available at a certain rank. If your tool provides a score for a given prediction it may be added, but the score fields may also be left empty. Please use the provided .csv template here.
- SequenceID (fasta header)
- Realm (-viria)
- Realm_score
- Subrealm (-vira)
- Subrealm_score
- Kingdom (-virae)
- Kingdom_score
- Subkingdom (-virites)
- Subkingdom_score
- Phylum (-viricota)
- Phylum_score
- Subphylum (-viricotina)
- Subphylum_score
- Class (-viricetes)
- Class_score
- Subclass (-viricetidae)
- Subclass_score
- Order (-virales)
- Order_score
- Suborder (-virineae)
- Suborder_score
- Family (-viridae)
- Family_score
- Subfamily (-virinae)
- Subfamily_score
- Genus (-virus)
- Genus_score
- Subgenus (-virus)
- Subgenus_score
- Species (binomial)
- Species_score
| SequenceID | Realm | Realm_score | Subrealm | Subrealm_score | Kingom | Kingom_score | ... | Genus | Genus_score | Subgenus | Subgenus_score | Species | Species_score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICTVTaxoChallenge_XXXXX | Varidnaviria | 0.77 | NA | NA | Bamfordvirae | 0.54 | ... | Mimivirus | 0.92 | NA | NA | Mimivirus lagoaense | 0.92 |
How can you send your results?
Please let us know by email where we can find your repository when you are done with the analysis. The repository should have a “results” folder where the predictions can be found as a .tsv file according to the template we provided. The README of your repository should provide the necessary instructions to easily reproduce the results using the code available in your repository. It should also contain a brief description of the methodology used for the classification. If you should prefer to keep your repository private until the end of the challenge, you can invite “0mician” and “dutilh” to your GitHub repository, but please note that the code should be made public by the time of the write up of the challenge’s report (see “what’s next”). If you would like to use another git repository or if repositories are not possible for you but you would still want to submit an entry, please contact us by email (see below).
What is next for the challenge
The deadline for submitting the results is December 31st 2024. We envision writing a report of the community’s finding and believe this will inform current efforts within the ICTV. To this end, the results will be used for a first draft of the manuscript to which all participants will be invited to respond and provide input.
Contact
Any questions or suggestions? Please feel free to contact Cédric Lood and Bas Dutilh, we will be happy to discuss with you. Our email addresses can be found here.
References
- Peter Simmonds, Evelien M. Adriaenssens, F. Murilo Zerbini, Nicola G.A. Abrescia, Pakorn Aiewsakun, Poliane Alfenas-Zerbini, Yiming Bao, Jakub Barylski, Christian Drosten, Siobain Duffy, W. Paul Duprex, Bas E. Dutilh, Santiago F. Elena, María Laura García, Sandra Junglen, Aris Katzourakis, Eugene V. Koonin, Mart Krupovic, Jens H. Kuhn, Amy J. Lambert, Elliot J. Lefkowitz, Małgorzata Łobocka, Cédric Lood, Jennifer Mahony, Jan P. Meier-Kolthoff, Arcady R. Mushegian, Hanna M. Oksanen, Minna M. Poranen, Alejandro Reyes-Muñoz, David L. Robertson, Simon Roux, Luisa Rubino, Sead Sabanadzovic, Stuart Siddell, Tim Skern, Donald B. Smith, Matthew B. Sullivan, Nobuhiro Suzuki, Dann Turner, Koenraad Van Doorslaer, Anne-Mieke Vandamme, Arvind Varsani, and Nikos Vasilakis (2023), Four principles to establish a universal virus taxonomy, PLoS Biology 21: e3001922, doi: 10.1371/journal.pbio.3001922